Using your our own glacier inventory with OGGM
Contents
Using your our own glacier inventory with OGGM#
The Randolph Glacier Inventory is a key dataset to model glaciers at any scale: it includes outlines of the extent of each glacier in the world, an information that is critical in figuring out how much a particular glacier might contribute to rising sea level. These glacier outlines are the starting point of any simulation in OGGM. The RGI’s latest version (v6), as well as v5, are provided and supported by OGGM (see the documentation). However, there are several issues in the RGI that might make you want to use your own corrected glacier outlines.
This notebook describes how to feed OGGM with them. We will show you three case studies about how to give any geometry to OGGM and avoid errors of incompatibility between your shapefile and the model framework.
We have three case studies which should cover a number of applications:
Dividing a glacier into smaller entities (common case, useful for poorly outlined glaciers, which are in reality separate dynamical entities)
Merging two glaciers together (useful for tidewater glaciers in particular, not much elsewhere)
TLDR;#
If you want to use custom data to feed OGGM with, you should:
make a shapefile that resembles the RGI one: same attributes, and the glacier geometries should be in lon/lat projection. The most important attribute is
geometry, of course, but others are used by OGGM as well: refer to the OGGM documentation to decide which ones. The RGI documentation (found in the RGI directory after download) is useful as well! We also have a useful function (oggm.utils.cook_rgidf) which can help you with that.compute and use a new glacier interesects file, or make sure you don’t need one and disable this option in OGGM.
Structure of an RGI file#
import numpy as np
import pandas as pd
import geopandas as gpd
import matplotlib.pyplot as plt
import oggm
import os
from oggm import cfg, utils, workflow, tasks, graphics
from oggm.core import inversion
cfg.initialize(logging_level='WARNING')
2023-03-07 12:47:00: oggm.cfg: Reading default parameters from the OGGM `params.cfg` configuration file.
2023-03-07 12:47:00: oggm.cfg: Multiprocessing switched OFF according to the parameter file.
2023-03-07 12:47:00: oggm.cfg: Multiprocessing: using all available processors (N=2)
Let’s read a file from the standard RGI:
utils.get_rgi_dir(version='61')
'/github/home/OGGM/rgi/RGIV61'
sh = utils.get_rgi_region_file('11', version='61')
sh
'/github/home/OGGM/rgi/RGIV61/11_rgi61_CentralEurope/11_rgi61_CentralEurope.shp'
Shapefiles are best read an manipulated with geopandas in python (see also our working_with_rgi.ipynb tutorial):
gdf = gpd.read_file(sh)
gdf.head()
| RGIId | GLIMSId | BgnDate | EndDate | CenLon | CenLat | O1Region | O2Region | Area | Zmin | ... | Lmax | Status | Connect | Form | TermType | Surging | Linkages | Name | check_geom | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | RGI60-11.00001 | G013599E47495N | 20030799 | 20030999 | 13.5987 | 47.4949 | 11 | 1 | 0.122 | 2191 | ... | 461 | 0 | 0 | 0 | 0 | 9 | 9 | NaN | NaN | POLYGON ((13.60035 47.49330, 13.59995 47.49332... |
| 1 | RGI60-11.00002 | G013614E47485N | 20030799 | 20030999 | 13.6135 | 47.4845 | 11 | 1 | 2.292 | 2203 | ... | 1853 | 0 | 0 | 0 | 0 | 9 | 9 | NaN | NaN | POLYGON ((13.60638 47.47578, 13.60599 47.47579... |
| 2 | RGI60-11.00003 | G013596E47484N | 20030799 | 20030999 | 13.5960 | 47.4835 | 11 | 1 | 0.851 | 2280 | ... | 1140 | 0 | 0 | 0 | 0 | 9 | 9 | NaN | NaN | POLYGON ((13.59765 47.47613, 13.59726 47.47614... |
| 3 | RGI60-11.00004 | G013583E47481N | 20030799 | 20030999 | 13.5829 | 47.4807 | 11 | 1 | 0.053 | 2319 | ... | 382 | 0 | 0 | 0 | 0 | 9 | 9 | NaN | NaN | POLYGON ((13.58283 47.47969, 13.58243 47.47971... |
| 4 | RGI60-11.00005 | G013603E47477N | 20030799 | 20030999 | 13.6026 | 47.4774 | 11 | 1 | 0.057 | 2656 | ... | 202 | 0 | 0 | 0 | 0 | 9 | 9 | NaN | NaN | POLYGON ((13.60076 47.47519, 13.60036 47.47521... |
5 rows × 24 columns
An RGI file contains the actual glacier geometries, but also a number of attribute which are used by OGGM afterwards. Let’s learn how to make our own file now.
Case 1: dividing a glacier into smaller entities#
A typical example of wrongly divided glacier is Hintereisferner, in Austria:
# OGGM set-up
cfg.PATHS['working_dir'] = utils.gettempdir(dirname='rgi-case-1', reset=True)
cfg.PARAMS['border'] = 10
# Get the HEF geometry and plot it
gl = utils.get_rgi_glacier_entities(['RGI60-11.00897'])
gl.plot(edgecolor='k');
2023-03-07 12:47:02: oggm.cfg: PARAMS['border'] changed from `40` to `10`.
Obviously, the two smaller tongues used to flow in the main one but this is not the case anymore today. We need updated geometries.
Make a new “RGI file”#
There is no simple way to automate the process of finding these bad geometries, but we are working on this (don’t hold your breath, this has been in development since a long time). Here we use a geometry that we prepared in QGis:
# We simulate the case where we only have the geometry, nothing else
divides = gpd.read_file(utils.get_demo_file('divides_alps.shp'))
divides = divides.loc[divides.RGIId == 'RGI50-11.00897'][['geometry']]
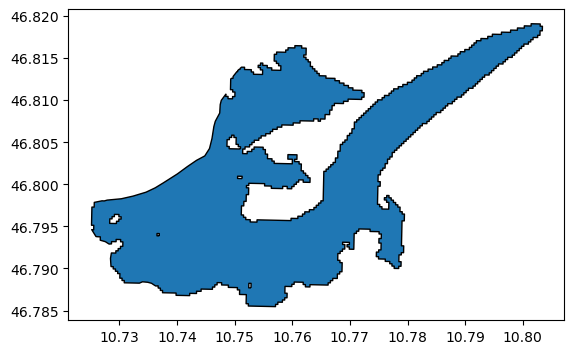
divides
| geometry | |
|---|---|
| 1 | POLYGON ((10.74505 46.80376, 10.74610 46.80293... |
| 93 | POLYGON ((10.74860 46.80493, 10.74717 46.80529... |
| 94 | POLYGON ((10.74602 46.80561, 10.74717 46.80529... |
divides.plot(edgecolor='k');
Now we use the RGI entity as template - it’s good to use the same attributes as the original RGI glacier, because most of them are already correct:
template = pd.concat([gl]*3, ignore_index=True)
template
| RGIId | GLIMSId | BgnDate | EndDate | CenLon | CenLat | O1Region | O2Region | Area | Zmin | ... | Aspect | Lmax | Status | Connect | Form | TermType | Surging | Linkages | Name | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | RGI60-11.00897 | G010758E46800N | 20030799 | 20030999 | 10.7584 | 46.8003 | 11 | 1 | 8.036 | 2430 | ... | 71 | 7178 | 0 | 0 | 0 | 0 | 9 | 1 | Hintereisferner | POLYGON ((10.75085 46.81381, 10.75112 46.81397... |
| 1 | RGI60-11.00897 | G010758E46800N | 20030799 | 20030999 | 10.7584 | 46.8003 | 11 | 1 | 8.036 | 2430 | ... | 71 | 7178 | 0 | 0 | 0 | 0 | 9 | 1 | Hintereisferner | POLYGON ((10.75085 46.81381, 10.75112 46.81397... |
| 2 | RGI60-11.00897 | G010758E46800N | 20030799 | 20030999 | 10.7584 | 46.8003 | 11 | 1 | 8.036 | 2430 | ... | 71 | 7178 | 0 | 0 | 0 | 0 | 9 | 1 | Hintereisferner | POLYGON ((10.75085 46.81381, 10.75112 46.81397... |
3 rows × 23 columns
We change the important ones:
# Attributes
template['RGIId'] = ['RGI60-11.00897_d01', 'RGI60-11.00897_d02', 'RGI60-11.00897_d03']
template['Name'] = ['Hintereisferner d01', 'Hintereisferner d02', 'Hintereisferner d03']
# Geometries
template['geometry'] = divides['geometry'].values
# Center point
for i, geom in template[['geometry']].iterrows():
cenlon, cenlat = geom.geometry.centroid.xy
template.loc[i, 'CenLon'] = np.array(cenlon)
template.loc[i, 'CenLat'] = np.array(cenlat)
# This is important to properly georeference the file
import salem
template.crs = salem.wgs84.srs
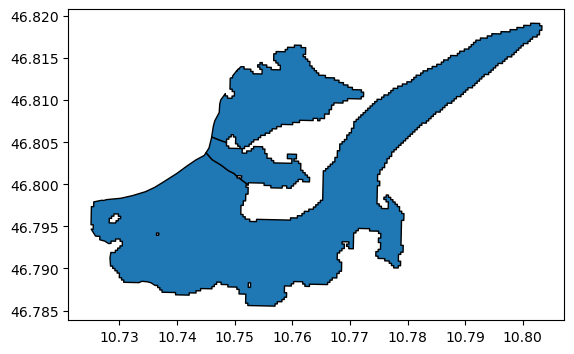
template.plot(edgecolor='k');
Save it:
hef_new_shape_path = os.path.join(cfg.PATHS['working_dir'], 'hef_divided.shp')
template.to_file(hef_new_shape_path)
Compute the intersects#
Hintereisferner has a divide with another glacier as well! Let’s find out which:
intersects_alps = gpd.read_file(utils.get_rgi_intersects_region_file('11'))
intersects_hef = intersects_alps.loc[(intersects_alps.RGIId_1 == 'RGI60-11.00897') | (intersects_alps.RGIId_2 == 'RGI60-11.00897')]
intersects_hef
| RGIId_1 | RGIId_2 | geometry | |
|---|---|---|---|
| 119 | RGI60-11.00846 | RGI60-11.00897 | LINESTRING (10.74826 46.81077, 10.74840 46.810... |
| 143 | RGI60-11.00897 | RGI60-11.00846 | LINESTRING (10.72557 46.79781, 10.72557 46.797... |
Ok, we can now create a file which has all the glaciers we need to compute the relevant intersects (note that we could also use the full standard RGI with just HEF replaced):
for_intersects = pd.concat([template, utils.get_rgi_glacier_entities(['RGI60-11.00846'])], ignore_index=True)
for_intersects.crs = salem.wgs84.srs
for_intersects.plot(edgecolor='k');
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[13], line 1
----> 1 for_intersects = pd.concat([template, utils.get_rgi_glacier_entities(['RGI60-11.00846'])], ignore_index=True)
2 for_intersects.crs = salem.wgs84.srs
3 for_intersects.plot(edgecolor='k');
File /usr/local/pyenv/versions/3.10.10/lib/python3.10/site-packages/pandas/util/_decorators.py:331, in deprecate_nonkeyword_arguments.<locals>.decorate.<locals>.wrapper(*args, **kwargs)
325 if len(args) > num_allow_args:
326 warnings.warn(
327 msg.format(arguments=_format_argument_list(allow_args)),
328 FutureWarning,
329 stacklevel=find_stack_level(),
330 )
--> 331 return func(*args, **kwargs)
File /usr/local/pyenv/versions/3.10.10/lib/python3.10/site-packages/pandas/core/reshape/concat.py:381, in concat(objs, axis, join, ignore_index, keys, levels, names, verify_integrity, sort, copy)
159 """
160 Concatenate pandas objects along a particular axis.
161
(...)
366 1 3 4
367 """
368 op = _Concatenator(
369 objs,
370 axis=axis,
(...)
378 sort=sort,
379 )
--> 381 return op.get_result()
File /usr/local/pyenv/versions/3.10.10/lib/python3.10/site-packages/pandas/core/reshape/concat.py:616, in _Concatenator.get_result(self)
612 indexers[ax] = obj_labels.get_indexer(new_labels)
614 mgrs_indexers.append((obj._mgr, indexers))
--> 616 new_data = concatenate_managers(
617 mgrs_indexers, self.new_axes, concat_axis=self.bm_axis, copy=self.copy
618 )
619 if not self.copy:
620 new_data._consolidate_inplace()
File /usr/local/pyenv/versions/3.10.10/lib/python3.10/site-packages/pandas/core/internals/concat.py:226, in concatenate_managers(mgrs_indexers, axes, concat_axis, copy)
223 values = np.concatenate(vals, axis=1)
224 else:
225 # TODO(EA2D): special-casing not needed with 2D EAs
--> 226 values = concat_compat(vals, axis=1)
227 values = ensure_block_shape(values, ndim=2)
229 values = ensure_wrapped_if_datetimelike(values)
File /usr/local/pyenv/versions/3.10.10/lib/python3.10/site-packages/pandas/core/dtypes/concat.py:133, in concat_compat(to_concat, axis, ea_compat_axis)
130 if isinstance(to_concat[0], ABCExtensionArray):
131 # TODO: what about EA-backed Index?
132 cls = type(to_concat[0])
--> 133 return cls._concat_same_type(to_concat)
134 else:
135 return np.concatenate(to_concat)
File /usr/local/pyenv/versions/3.10.10/lib/python3.10/site-packages/geopandas/array.py:1323, in GeometryArray._concat_same_type(cls, to_concat)
1311 """
1312 Concatenate multiple array
1313
(...)
1320 ExtensionArray
1321 """
1322 data = np.concatenate([ga.data for ga in to_concat])
-> 1323 return GeometryArray(data, crs=_get_common_crs(to_concat))
File /usr/local/pyenv/versions/3.10.10/lib/python3.10/site-packages/geopandas/array.py:1413, in _get_common_crs(arr_seq)
1406 warnings.warn(
1407 "CRS not set for some of the concatenation inputs. "
1408 f"Setting output's CRS as {names[0]} "
1409 "(the single non-null crs provided)."
1410 )
1411 return crs_not_none[0]
-> 1413 raise ValueError(
1414 f"Cannot determine common CRS for concatenation inputs, got {names}. "
1415 "Use `to_crs()` to transform geometries to the same CRS before merging."
1416 )
ValueError: Cannot determine common CRS for concatenation inputs, got ['unknown', 'WGS 84']. Use `to_crs()` to transform geometries to the same CRS before merging.
Good! Let’s use rgitools to compute the intersects for this new situation:
from rgitools.funcs import compute_intersects
new_intersects = compute_intersects(for_intersects)
If this raises an error, you might have to install rgitools first (e.g. pip install git+https://github.com/OGGM/rgitools.git)
and also networkx (pip install networkx)
f, ax = plt.subplots()
for_intersects.plot(ax=ax, edgecolor='k');
new_intersects.plot(ax=ax, edgecolor='r');
Good! We can store our intersects to use them with OGGM afterwards:
hef_intersects_path = os.path.join(cfg.PATHS['working_dir'], 'hef_divided_intersects.shp')
new_intersects.to_file(hef_intersects_path)
Finally: the OGGM run#
# This is important! We tell OGGM to recompute the glacier area for us
cfg.PARAMS['use_rgi_area'] = False
# This is the default anyway, but we set it here to be sure
cfg.PARAMS['use_intersects'] = True
# This is important!
cfg.set_intersects_db(hef_intersects_path)
# This is to avoid a download in the tutorial, you dont' need do this at home
cfg.PATHS['dem_file'] = utils.get_demo_file('hef_srtm.tif')
# This is important again - standard OGGM
rgidf = gpd.read_file(hef_new_shape_path)
gdirs = workflow.init_glacier_directories(rgidf, reset=True, force=True)
workflow.execute_entity_task(tasks.define_glacier_region, gdirs);
workflow.execute_entity_task(tasks.glacier_masks, gdirs);
workflow.execute_entity_task(tasks.compute_centerlines, gdirs);
workflow.execute_entity_task(tasks.initialize_flowlines, gdirs);
workflow.execute_entity_task(tasks.catchment_area, gdirs);
workflow.execute_entity_task(tasks.catchment_width_geom, gdirs);
workflow.execute_entity_task(tasks.catchment_width_correction, gdirs);
If you get “NameError ‘skdraw’ is not defined”, you have to install the python image processing package scikit-image first (e.g. via conda install scikit-image)
graphics.plot_catchment_width(gdirs, add_intersects=True, corrected=True)
It works!
The intersects in OGGM are used for two main things:
when a grid-point glacier section touches an intersect, it will be attributed a rectangular bed (instead of a parabolic one)
when interpolating the ice thickness to a 2D grid, the boundary condition thickness=0 at the glacier outline is removed where there are intersects
We recommend to use intersects for your runs as well.
Case 2: merging glaciers#
Sometimes, you may want to merge glaciers together. This case is less frequent than Case 1, but might be useful for calving glaciers, which are sometimes divided in the RGI.
Original RGI outlines#
We use a case study for two marine-terminating glaciers in Alaska that have to be merged into a single outline in order to model a correct calving flux for these glaciers (following the methods described in Recinos et al., (2019)). The resulting shapefile is a new one that needs to be adapted in order for OGGM to run.
We will study the Sawyer Glacier (RGI60-01.03890) that is actually connected via the calving front with this other entity (RGI60-01.23664). Visit this link to learn more about the retreat of the Sawyer Glacier and see images illustrating this connection.
gl = utils.get_rgi_glacier_entities(['RGI60-01.03890', 'RGI60-01.23664'])
Here OGGM downloaded the outlines for both glaciers. If we plot them together, we can see that both glaciers drain into the same fjord. See the google map below:
cfg.initialize(logging_level='WARNING')
cfg.PATHS['working_dir'] = utils.gettempdir(dirname='rgi-case-2-example', reset=True)
cfg.PARAMS['border'] = 10
gdirs = workflow.init_glacier_directories(['RGI60-01.03890', 'RGI60-01.23664'], from_prepro_level=3, reset=True, force=True)
graphics.plot_googlemap(gdirs, figsize=(6, 6));
The upper glacier map is a zoom version of the plot below. They share the same glaciers terminus. Therefore, to estimate a calving flux for these glaciers we need them connected.
Let’s merge these two outlines using geopandas#
merged = gl.dissolve(by='O2Region', as_index=False)
merged = merged[gl.columns]
merged.plot(edgecolor='k');
RGI attributes#
We now have a new shapefile, which resembles an RGI one but has wrong attributes. Some aren’t relevant, but some are. See the documentation for a list.
The important ones are: RGIId, CenLon, CenLat, TermType, Area. Area and CenLon, Cenlat can be calculated by OGGM later, as we have seen earlier. Here, we prefer to keep the Area computed by the RGI for consistency.
# We use the RGI as template (this avoids strange IO issues)
template = gl.iloc[[0]].copy()
template['geometry'] = merged['geometry'].values
# Change CenLon, CenLat
cenlon, cenlat = merged.iloc[0].geometry.representative_point().xy
template['CenLon'] = cenlon
template['CenLat'] = cenlat
# We sum up the areas
template['Area'] = gl.Area.sum()
template['Area']
In Recinos et al., (2019) we wanted to estimate a frontal ablation flux for this new outline and compare it with previous estimates found in the literature for the Sawyer Glacier (McNabb et al., 2015).
For this reason we kept the Sawyer glacier attributes to the following variables:
RGIId, GLIMSId, Name
The TermType should be equal to 1, for Marine-terminating.
template['TermType'] = 1
template['Name'] = 'Sawyer Glacier merged with RGI60-01.23664'
Now we can write a new shapefile for this glacier. We recommend doing this if you have to make several model runs. You can also integrate this outline to your main RGI shapefile:
cfg.initialize()
cfg.PATHS['working_dir'] = utils.gettempdir(dirname='rgi-case-2', reset=True)
template.crs = salem.wgs84.srs
template.to_file(os.path.join(cfg.PATHS['working_dir'], 'merged.shp'))
Run OGGM with this new glacier#
For simplicity, we do not compute the intersects in this case: however, we recommend you do do so (see above). In all cases, do not use the intersects provided automatically with OGGM when using custom inventories, as they are likely to be wrong.
# Set-up the run
cfg.PARAMS['border'] = 10
# We don't use intersects here
cfg.PARAMS['use_intersects'] = False
# We prefer OGGM to use the area we computed ourselves
cfg.PARAMS['use_rgi_area'] = True
# Use our merged file
rgidf = gpd.read_file(os.path.join(cfg.PATHS['working_dir'], 'merged.shp'))
gdirs = workflow.init_glacier_directories(rgidf, reset=True, force=True)
gdirs
Here we are not able to use the Pre-processed directories and the respective Processing levels that OGGM provides for a easy run set up. We can’t use this workflow simply because we have a different beginning than OGGM, we have a different RGI! We just need to type more and run all the model task one by one:
from oggm.workflow import execute_entity_task
# Calculate the DEMs and the masks
execute_entity_task(tasks.define_glacier_region, gdirs, source = 'SRTM')
execute_entity_task(tasks.glacier_masks, gdirs)
# Calculate the Pre-processing tasks
task_list = [
tasks.compute_centerlines,
tasks.initialize_flowlines,
tasks.catchment_area,
tasks.catchment_width_geom,
tasks.catchment_width_correction,
]
for task in task_list:
execute_entity_task(task, gdirs)
note that we used in tasks.define_glacier_region SRTM as DEM source which is not the default one in OGGM.
The default one is NASADEM, but for that you have to register at NASA Earthdata
more info in dem_sources.ipynb/register
graphics.plot_googlemap(gdirs, figsize=(6, 6));
graphics.plot_catchment_width(gdirs, corrected=True);
Case 3: start from a completely independent inventory#
OGGM (since version 1.5.3) now offers a function (utils.cook_rgidf()) to make it easier of using a non-RGI glacier inventory in OGGM. Now, let’s use a non-RGI glacier inventory from the second Chinese glacier inventory (CGI2, https://doi.org/10.3189/2015JoG14J209) to show how it works.
# Let's get the sample CGI2 glacier inventory and see what it looks like
from oggm import utils
import geopandas as gpd
cgidf = gpd.read_file(utils.get_demo_file('cgi2.shp'))
cgidf
Ha! There are some Chinese characters! But it should not influence our work. Now, let’s try with the simplest case:
rgidf_simple = utils.cook_rgidf(cgidf, o1_region='13')
rgidf_simple
In this case, we fake all of the columns values except for geometry. With this rgidf_simple, we can handle most of the OGGM procedure after set cfg.PARAMS['use_rgi_area'] = False. Let’s have a try:
from oggm import cfg, workflow
cfg.initialize()
cfg.PARAMS['use_multiprocessing'] = True
cfg.PARAMS['use_rgi_area'] = False
cfg.PARAMS['use_intersects'] = False
cfg.PATHS['working_dir'] = utils.gettempdir(dirname='cook_rgidf', reset=True)
gdirs = workflow.init_glacier_directories(rgidf_simple)
# The tasks below require downloading new data - we comment them for the tutorial, but it should work for you!
# workflow.gis_prepro_tasks(gdirs)
# workflow.download_ref_tstars('https://cluster.klima.uni-bremen.de/~oggm/ref_mb_params/oggm_v1.4/RGIV62/CRU/centerlines/qc3/pcp2.5')
# workflow.climate_tasks(gdirs)
# workflow.inversion_tasks(gdirs)
utils.compile_glacier_statistics(gdirs)
Sometimes, the information in the original glacier inventory also covered what RGI need, but with different name. For example, both CGI2 and RGI include Lon-lat coordinate to indicate the location of the glacier but with different names. CGI2 used Glc_Long and Glc_Lati, while RGI6 used CenLon and CenLat. Also, CGI2 include glacier area information with name Glc_Area, while RGI named glacier area as Area. If we want to keep those values but rename them following RGI, we can use the assign_column_values keyword argument:
rgidf_save_columns = utils.cook_rgidf(cgidf, o1_region='13', assign_column_values={'Glc_Long': 'CenLon', 'Glc_Lati': 'CenLat', 'Glc_Area': 'Area', 'GLIMS_ID': 'GLIMSId'})
rgidf_save_columns
Seems perfect! However, the glacier area in CGI2 is in \(m^2\), but it is in \(km^2\) for RGI. So, we need to correct the Area values right:
rgidf_save_columns['Area'] = rgidf_save_columns.Area * 1e-6
rgidf_save_columns.Area
Note:#
Despite that cook_rgidf() can handle most of the cases for OGGM, there are some limitations. Here, we try to point out some of cases in which the rgidf sourced from cook_rgidf() might get you in trouble:
in
cook_rgidf(), we assign the glacier form with ‘0’ (Glacier) for all of the glaciers in the original data. OGGM assigns different parameters for glaciers (form ‘0’) and ice caps (form ‘1’).termtypewas also assign as ‘0’ which means ‘land-terminating’. Here again, OGGM treats ‘Marine-terminating’ glaciers differently (see ‘Frontal ablation’).
For these kinds of attribution, there is nothing we can do automatically. Users need to assign the right values according the actual condition of their glaciers, if the attribution is important to their use cases.
What’s next?#
return to the OGGM documentation
back to the table of contents


