10 minutes to… a preprocessed directory#
This tutorial provides a first introduction to running OGGM. The OGGM workflow is best understood through a concrete example, and in the following we will guide you through the core concepts and workflow of OGGM (model structure and fundamentals).
This example is also designed to help you set up OGGM for the first time on your own computer. If you prefer not to install OGGM locally, you can run this notebook online instead via MyBinder or OGGM-Hub.
Tags: beginner, glacier-directory, workflow
First time set-up#
Input data folders#
If you are using your own computer: before you start, make sure that you have set-up the input data configuration file at your wish.
In the course of this tutorial, we will need to download data needed for each glacier (a few 100s of mb at max, depending on the chosen glaciers), so make sure you have an internet connection.
cfg.initialize() and cfg.PARAMS#
An OGGM simulation script will always start with the following commands:
from oggm import cfg, utils
cfg.initialize()
2026-03-09 23:08:53: oggm.cfg: Reading default parameters from the OGGM `params.cfg` configuration file.
2026-03-09 23:08:53: oggm.cfg: Multiprocessing switched OFF according to the parameter file.
2026-03-09 23:08:53: oggm.cfg: Multiprocessing: using all available processors (N=4)
A call to cfg.initialize() will read the default parameter file (or any user-provided file) and make them available to all other OGGM tools via the cfg.PARAMS dictionary. Here are some examples of these parameters:
cfg.PARAMS['melt_f'], cfg.PARAMS['ice_density'], cfg.PARAMS['continue_on_error']
(5.0, 900.0, False)
See here for the default parameter file and a description of their role and default value.
# You can try with or without multiprocessing: with two glaciers, OGGM could run on two processors
cfg.PARAMS['use_multiprocessing'] = True
2026-03-09 23:08:54: oggm.cfg: Multiprocessing switched ON after user settings.
Workflow#
In this section, we will explain the fundamental concepts of the OGGM workflow:
Working directories
Glacier directories
Tasks
Working directory#
Each OGGM run needs a single folder where to store the results of the computations for all glaciers. This is called a “working directory” and needs to be specified before each run. Here we create a temporary folder for you:
cfg.PATHS['working_dir'] = utils.gettempdir(dirname='OGGM-GettingStarted-10m', reset=True)
cfg.PATHS['working_dir']
'/tmp/OGGM/OGGM-GettingStarted-10m'
We use a temporary directory for this example (a directory which will be deleted by your operating system next time to restart your computer), but in practice for a real simulation, you will set this working directory yourself (for example: /home/zoe/OGGM_output). The size of this directory will depend on how many glaciers you’ll simulate!
You can create a persistent OGGM working directory at a specific path via path = utils.mkdir(path). Beware! If you use reset=True in utils.mkdir, ALL DATA in this folder will be deleted!
Define the glaciers for the run#
rgi_ids = ['RGI60-11.01328', 'RGI60-11.01450']
You can provide any number of glacier identifiers to OGGM. In this case, we chose:
RGI60-11.01328: Unteraar Glacier in the Swiss AlpsRGI60-11.01450: Aletsch Glacier in the Swiss Alps
Here is a list of other glaciers you might want to try out:
RGI60-11.00897: Hintereisferner in the Austrian Alps.RGI60-18.02342: Tasman Glacier in New ZealandRGI60-11.00787: Kesselwandferner in the Austrian Alps… or any other glacier identifier! You can find other glacier identifiers by exploring the GLIMS viewer. See the working with the RGI tutorial for an introduction on RGI IDs and the GLIMS browser.
For an operational run on an RGI region, you might want to download the Randolph Glacier Inventory dataset instead, and start a run from it. This case is covered in the working with the RGI tutorial.
Glacier directories#
The OGGM workflow is organized as a list of tasks that have to be applied to a list of glaciers. The vast majority of tasks are called entity tasks: they are standalone operations to be realized on one single glacier entity. These tasks are executed sequentially (one after another): they often need input generated by the previous task(s): for example, the climate calibration needs the glacier flowlines, which can be only computed after the topography data has been processed, and so on.
To handle this situation, OGGM uses a workflow based on data persistence on disk: instead of passing data as python variables from one task to another, each task will read the data from disk and then write the computation results back to the disk, making these new data available for the next task in the queue. These glacier specific data are located in glacier directories.
One main advantage of this workflow is that OGGM can prepare data and make it available to everyone! Here is an example of a url where such data can be found:
from oggm import workflow
from oggm import DEFAULT_BASE_URL
DEFAULT_BASE_URL
'https://cluster.klima.uni-bremen.de/~oggm/gdirs/oggm_v1.6/L3-L5_files/2025.6/elev_bands/W5E5/per_glacier_spinup/'
Let’s use OGGM to download the glacier directories for our two selected glaciers:
gdirs = workflow.init_glacier_directories(
rgi_ids, # which glaciers?
prepro_base_url=DEFAULT_BASE_URL, # where to fetch the data?
from_prepro_level=4, # what kind of data?
prepro_border=80 # how big of a map?
)
2026-03-09 23:08:54: oggm.workflow: init_glacier_directories from prepro level 4 on 2 glaciers.
2026-03-09 23:08:54: oggm.workflow: Execute entity tasks [gdir_from_prepro] on 2 glaciers
the keyword
from_prepro_levelindicates that we will start from pre-processed directories, i.e. data that are already prepared by the OGGM team. In many cases you will want to start from pre-processed directories, in most cases from level 3 or 5. For level 3 and above the model has already been calibrated, so you no longer need to do that yourself and can start right away with your simulation. Here we start from level 4 and add some data to the processing in order to demonstrate the OGGM workflow.the
prepro_borderkeyword indicates the number of grid points which we’d like to add to each side of the glacier for the local map: the larger the glacier will grow, the larger the border parameter should be. The available pre-processed border values are: 10, 80, 160, 240 (depending on the model set-ups there might be more or less options). These are the fixed map sizes we prepared for you - any other map size will require a full processing (see the further DEM sources example for a tutorial).
The init_glacier_directories task will allways be the very first task to call for all your OGGM experiments. Let’s see what it gives us back:
type(gdirs), type(gdirs[0])
(list, oggm.utils._workflow.GlacierDirectory)
gdirs is a list of GlacierDirectory objects (one for each glacier). Glacier directories are used by OGGM as “file and attribute manager” for single glaciers. For example, the model now knows where to find the topography data file for this glacier:
gdir = gdirs[0] # take Unteraar glacier
print('Path to the DEM:', gdir.get_filepath('dem'))
Path to the DEM: /tmp/OGGM/OGGM-GettingStarted-10m/per_glacier/RGI60-11/RGI60-11.01/RGI60-11.01328/dem.tif
And we can also access some attributes of this glacier:
gdir
<oggm.GlacierDirectory>
RGI id: RGI60-11.01328
Region: 11: Central Europe
Subregion: 11-01: Alps
Glacier type: Glacier
Terminus type: Land-terminating
Status: Glacier or ice cap
Area: 23.825 km2
Lon, Lat: (8.2193, 46.5642)
Grid (nx, ny): (299, 274)
Grid (dx, dy): (78.0, -78.0)
gdir.rgi_date # date at which the outlines are valid
2003
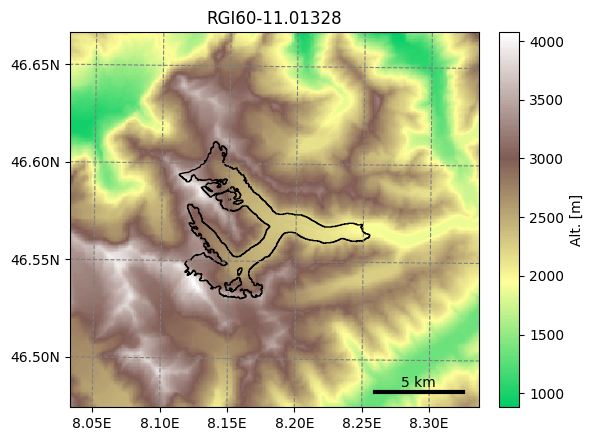
The advantage of this Glacier Directory data model is that it simplifies greatly the data transfer between tasks. The single mandatory argument of most OGGM commands will allways be a glacier directory. With the glacier directory, each OGGM task will find the input it needs: for example, both the glacier’s topography and outlines are needed for the next plotting function, and both are available via the gdir argument:
from oggm import graphics
graphics.plot_domain(gdir, figsize=(6, 5))
Another advantage of glacier directories is their persistence on disk: once created, they can be recovered from the same location by using init_glacier_directories again, but without keyword arguments:
# Fetch the LOCAL pre-processed directories - note that no arguments are used!
gdirs = workflow.init_glacier_directories(rgi_ids)
2026-03-09 23:08:55: oggm.workflow: Execute entity tasks [GlacierDirectory] on 2 glaciers
See the store_and_compress_glacierdirs tutorial for more information on glacier directories and how to use them for operational workflows.
Accessing data in the preprocessed directories#
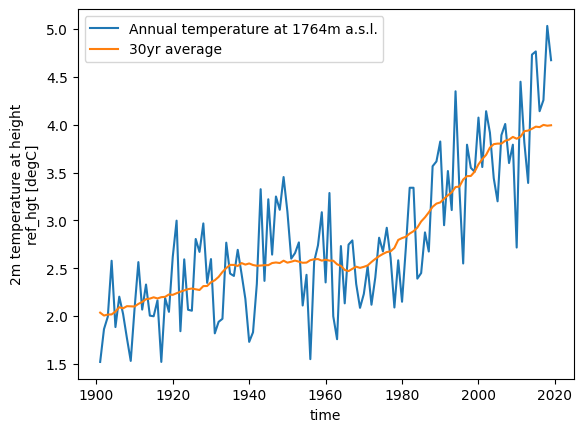
Glacier directories are the central object for model users and developers to access data for this glacier. Let’s say for example that you would like to retrieve the climate data that we have prepared for you. You can ask the glacier directory to tell you where this data is:
gdir.get_filepath('climate_historical')
'/tmp/OGGM/OGGM-GettingStarted-10m/per_glacier/RGI60-11/RGI60-11.01/RGI60-11.01328/climate_historical.nc'
One can then use familiar tools to read and process the data further:
import xarray as xr
import matplotlib.pyplot as plt
# Open the file using xarray
with xr.open_dataset(gdir.get_filepath('climate_historical')) as ds:
ds = ds.load()
# Plot the data
ds.temp.resample(time='YS').mean().plot(label=f'Annual temperature at {int(ds.ref_hgt)}m a.s.l.')
ds.temp.resample(time='YS').mean().rolling(time=31, center=True, min_periods=15).mean().plot(label='30yr average')
plt.legend();
As a result of the processing workflow the glacier directories can store many more files. If you are interested, you can have a look at the list:
import os
print(os.listdir(gdir.dir))
['elevation_band_flowline.csv', 'glacier_grid.json', 'diagnostics.json', 'model_geometry_spinup_historical.nc', 'model_diagnostics_historical.nc', 'inversion_flowlines.pkl', 'dem_source.txt', 'inversion_output.pkl', 'mb_calib.json', 'intersects.tar.gz', 'model_flowlines_dyn_melt_f_calib.pkl', 'gridded_data.nc', 'inversion_input.pkl', 'outlines.tar.gz', 'dem.tif', 'climate_historical.nc', 'log.txt', 'fl_diagnostics_historical.nc', 'fl_diagnostics_spinup_historical.nc', 'model_geometry_historical.nc', 'model_diagnostics_spinup_historical.nc', 'model_flowlines.pkl', 'downstream_line.pkl']
For a short explanation of what these files are, see the glacier directory documentation. In practice, however, you will only access a couple of these files yourself.
OGGM tasks#
There are two different types of “tasks” in OGGM:
Entity Tasks : Standalone operations to be realized on one single glacier entity, independently of the others. The majority of OGGM tasks are entity tasks. They are parallelisable: the same task can run on several glaciers in parallel.
Global Tasks : Tasks which require to work on several glacier entities at the same time. Model parameter calibration or the compilation of several glaciers’ output are examples of global tasks.
OGGM implements a simple mechanism to run a specific task on a list of GlacierDirectory objects. Here, for the sake of the demonstration of how tasks work, we are going to compute something new from the directory:
from oggm import tasks
# run the compute_inversion_velocities task on all gdirs
workflow.execute_entity_task(tasks.compute_inversion_velocities, gdirs);
2026-03-09 23:08:55: oggm.workflow: Execute entity tasks [compute_inversion_velocities] on 2 glaciers
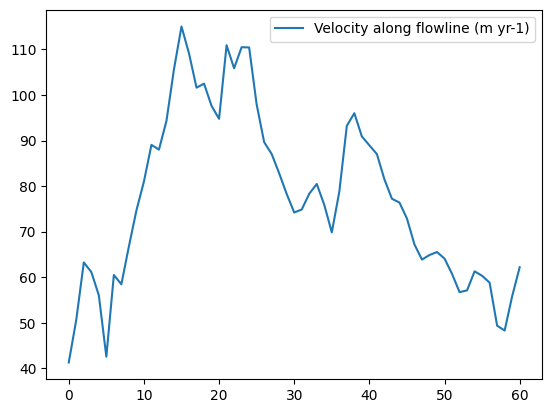
Compute inversion velocities is an optional task in the OGGM workflow. It computes the ice velocity along the flowline resulting from the ice flux from a glacier in equilibrium (more on this in the documentation or the dedicated tutorial).
Note that OGGM tasks often do not return anything (the statement above seems to be “void”, i.e. doing nothing). The vast majority of OGGM tasks actually write data to disk in order to retrieve it later. Let’s have a look at the data we just added to the directory:
inversion_output = gdir.read_pickle('inversion_output') # The task above wrote the data to a pickle file - but we write to plenty of other files!
# Take the first flowline
fl = inversion_output[0]
# the last grid points often have irrealistic velocities
# because of the equilibrium assumption
vel = fl['u_surface'][:-1]
plt.plot(vel, label='Velocity along flowline (m yr-1)'); plt.legend();
Recap#
OGGM is a “glacier centric” model, i.e. it operates on a list of glaciers.
OGGM relies on saving files to disk during the workflow. Therefore, users must always specify a working directory while running OGGM.
The working directory can be used to restart a run at a later stage.
Put simply, this “restart workflow” is what OGGM uses to deliver data to its users. Pre-processed directories are online folders filled with glacier data that users can download.
Once initialized locally, “glacier directories” allow OGGM to do what it does best: apply processing tasks to a list of glacier entities. These “entity tasks” get the data they need from disk, and write to disk once completed.
Users (or subsequent OGGM tasks) can use these data for new computations.
What’s next?#
visit the next tutorial: 10 minutes to… a glacier change projection with GCM data
do you want to understand how the preprocessed directories are built? Check out this step-by-step guide
back to the table of contents
return to the OGGM documentation


